Travel, Tech, Thoughts…
-
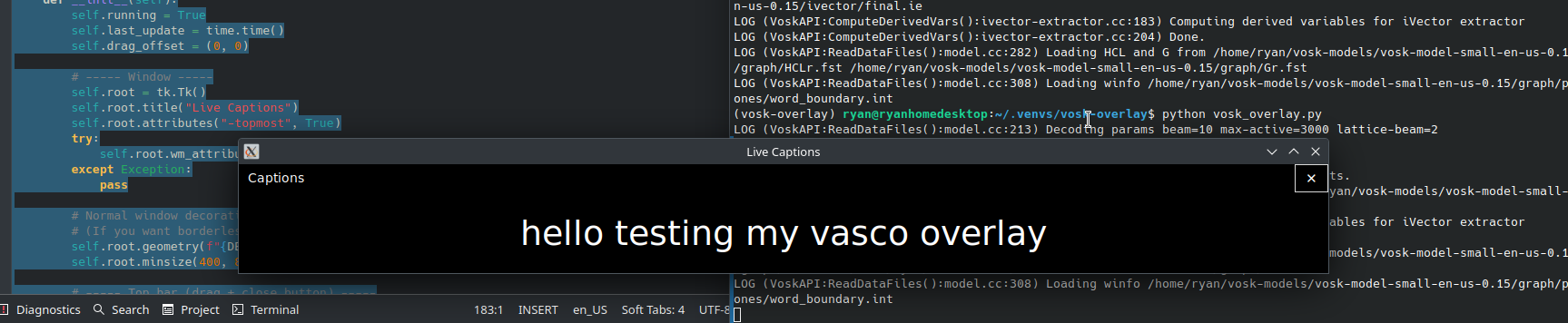
Voice-to-text Live Transcription on Kubuntu
(I’m running Kubuntu 24.04 with Plasma 5.27.12 and X11.) Here’s the situation. I’ve been told by my university administration that, within the next year, if I’m using a presentation (e.g., LibreOffice Impress) in my classes, I will have to have a voice-to-text option showing what I say as I talk to comply with federal requirements.…
-
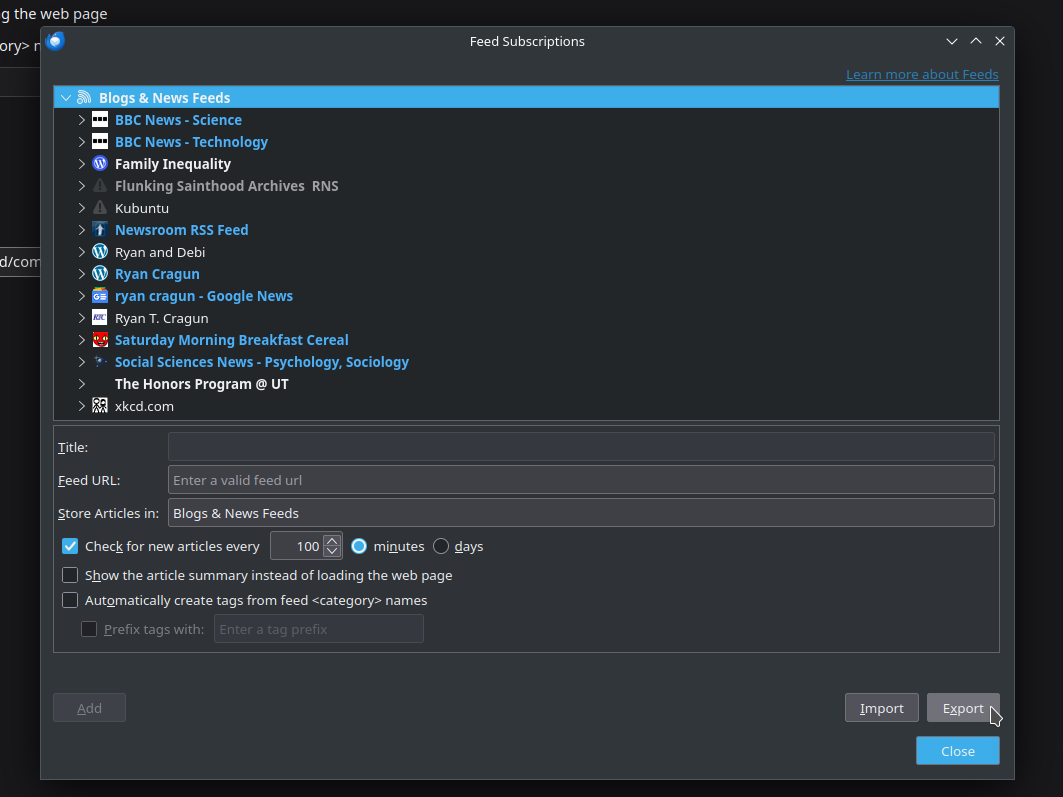
Thunderbird – Transfer RSS feeds
Thunderbird version: 140.6.0esr on Kubuntu 24.04 I use three different computers. I also use RSS feeds to get some of my news. For a while, I was paying for an online service that managed my RSS feeds. But I found I wasn’t accessing it enough to justify the cost. Since I have switched to Thunderbird…
-
LibreOffice Impress – Change Default Draw Color
(NOTE: The tutorial below is from LibreOffice 24.2.7.2) I have had an issue for a while (since I created my own custom color palette), and I finally found the solution. LibreOffice Impress comes with a standard template with default colors. As a college professor and researcher, I create lots of presentations. Since LibreOffice is my…
-
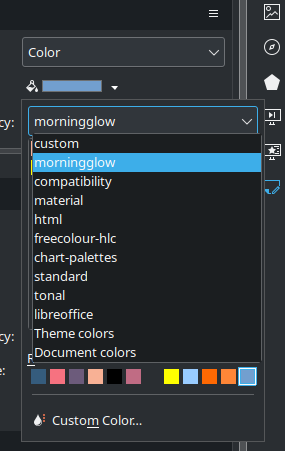
LibreOffice – Create Custom Color Palette
(This is for LibreOffice version 24.2.7.2 on Kubuntu 24.04.) I know I’m weird in a lot of ways (I mean, who still blogs in 2025?!?), but here’s a new one. I create lots of charts and graphs as well as presentations for classes. I tend to prefer a minimalist approach – white background with black…
-
AppImage, Snap, and Flatpak on Kubuntu
I’m guessing most people who don’t use Linux have no idea how convenient repositories are. I’m a huge fan of how Linux has managed (and mostly continues to manage) software for quite a while: you open a console or the software manager on your computer, search for the software you want, and install it. The…
-
Kubuntu 25.04 – KRunner and iBus issues
Over the last month, I have updated the three computers I use to Kubuntu 25.04. On my laptop, I ran into an issue where Wayland would just periodically freeze. I figured out a solution to that and detailed it here. I finally installed Kubuntu 25.04 on my home desktop, which is the machine I use…
-

Estonia – Suur Munamägi
Summit Date: 2025-07-08 Summit Party: Ryan Cragun and Thom Pittz This was the last highpoint on my quick tour through three Baltic countries – Lithuania, Latvia, and Estonia. We left Riga early that morning, stopped at the highest point in Latvia, then headed north into Estonia. It was a couple of hours between the two…
-

Latvia – Gaiziņkalns
Summit Date: 2025-07-08 Summit Party: Ryan Cragun and Thom Pittz As part of a short visit to the Baltic countries, I visited the highest points in Lithuania, Latvia, and Estonia. This is my brief high point report for Gaiziņkalns, the highest point in elevation in Latvia. I spent the previous day in Riga. A friend…
-

Lithuania – Aukštojas Hill
Date: 2025-07-05 Summit Party: Ryan Cragun and Thom Pittz I never would have thought I’d find myself in Lithuania, but a conference I regularly attend for work was held in Kaunas. A friend wanted to tour around a bit after the conference in the Baltics. Because it’s what I do, I immediately looked up the…
-

Convert Text to Audio on Linux – Coqui TTS
In a separate post on my blog, I documented a simple way to convert text to audio using Google’s Text-To-Speech service. This works for small projects. But, during a recent conversion, I ran into an error where my requests were getting blocked by Google for using the service too much. I didn’t realize they capped…