Tag: R
-
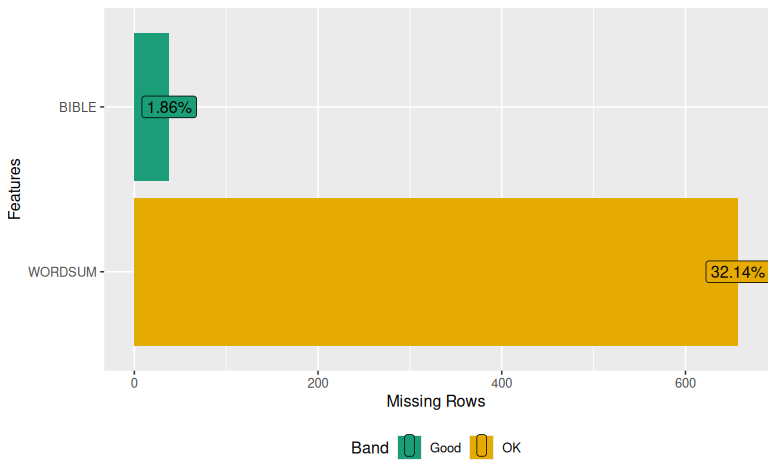
Imputing Missing Data in R – mice
I got a revise and resubmit on a recent paper. We noted that we had some missing data on some of the variables in our dataset. One of the reviewers suggested we address the missing data issue, which I have typically just managed using listwise/casewise deletion (a.k.a. complete-case analysis, which entails not including cases that…
-

R – Combining Data from Two Variables
(NOTE: This was done in R 4.3.1 using RStudio 2023.03.0.) Here’s the scenario: I have a variable (V1) from Country A with roughly ~1,000 responses that measures religious affiliation with the following options: To be clear, there are no responses to V1 from the participants in the other country (Country B), only from individuals from…
-

ChatGPT – checking for personally identifying information
I recently hired a panel survey company to conduct surveys in several countries around the world. With some of the questions, we gave participants the option to select “other” and then fill in the blank. Given the ethics approval that we have for the data, we are not allowed to retain any data that could…
-
R – importing data using haven
Introduction I hate to admit that I’ve been using R exclusively for data analysis for about 5 or 6 years and am just now realizing that I likely have been loading data sets into R incorrectly this entire time. Let me explain the issue. I regularly load datasets into R that are either in SPSS…
-
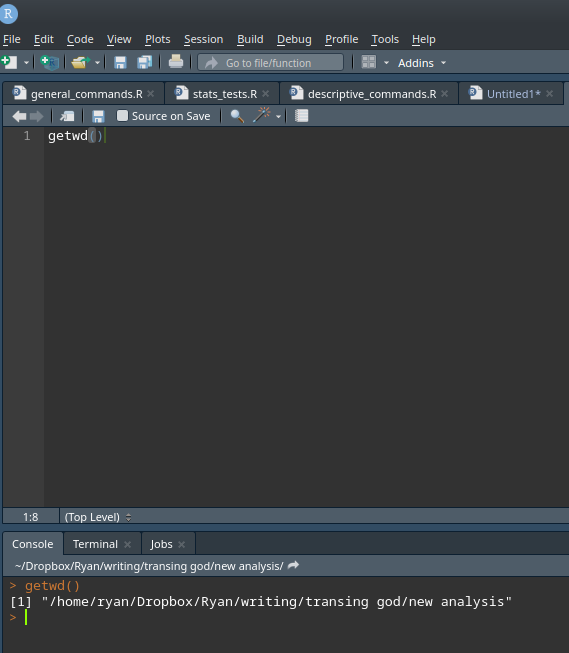
R – Finding and Setting Working Directory
R, like many software programs, likes to have a folder or directory on your computer to operate in. When you start R, depending on how you start it and whether or not you previously saved your session, it’s likely that R will set a default “working directory” or folder where it is going to look…
-
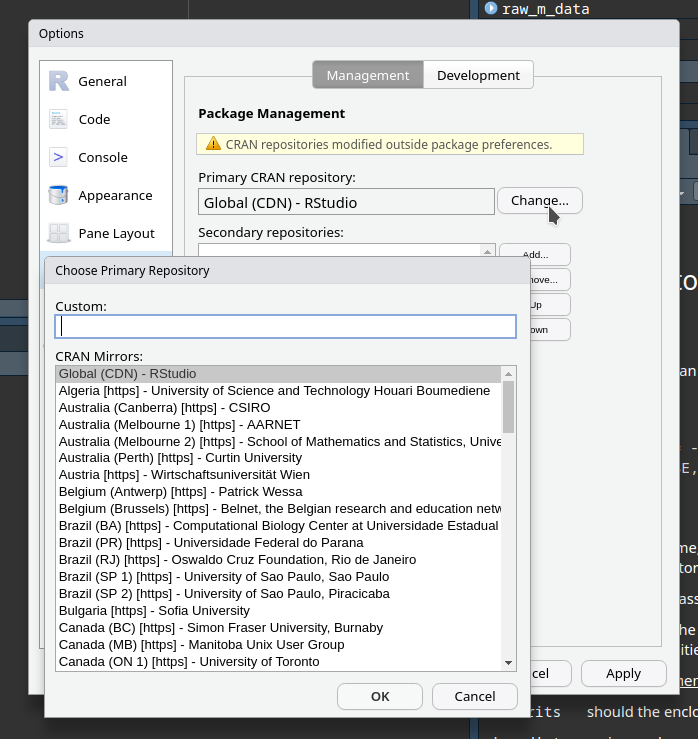
R – changing mirror in RStudio
CRAN (The Comprehensive R Archive Network) is a distributed set of servers that allow people to download the various R packages from a bunch of servers that are all “mirrors” of each other. These servers or mirrors are called “repositories.” There are many advantages to this: (1) people can download from a mirror that is…
-
R – create scatterplot with ggplot2
R has pretty amazing capabilities for creating charts and graphs. One of the most common packages for this is ggplot2. However, it’s not the most intuitive package I have used in R. So, I figured I’d illustrate how to make some relatively simple scatterplots in R using ggplot2. I’ll likely post instructions on how to…
-
R – find cases (rows) that match specific criteria
I regularly need to find a specific case or set of cases that meet some criteria when analyzing data, often so I can modify those values for one reason or another. The easiest way I have found to find such values in R is the “which” function. As with most of my R examples, I’m…
-
R – delete one or several variables in a dataset
I regularly create variables while analyzing data and then find that I need to delete a variable I created. At times, I just want to get rid of a variable in a dataset (’cause screw that variable). This short tutorial will explain how to delete a variable (or multiple variables if needed). As with most…
-
R – create variable filled with zeros
I ran into a situation where I needed to add a variable to a dataset. I knew that I was then going to modify some of the values in the variable, but most of the values were going to be zeros. So, I wanted to create a new variable and fill it with all zeros.…